research overview
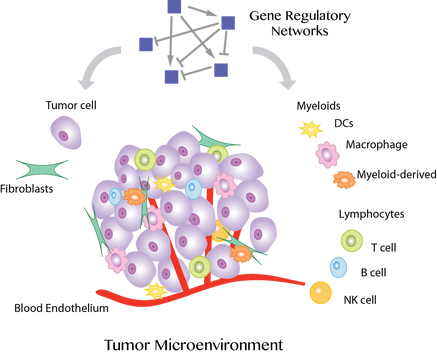
Cancer treatments lead to favorable outcomes only in a subset of patients partly due to significant heterogeneity in tumor cells. Towards developing improved and personalized treatments, our research aims to leverage machine learning to characterize the heterogeneous populations of interacting cells in the tumor microenvironment and their spatio-temporal dynamics and dysregulated circuitry.
Recent advances in single-cell and spatial genomic technologies have empowered us to profile heterogeneous phenotypes at the resolution of individual cells and in the context of the tissue microenvironment. However, analysis of high-dimensional genomic data involves major statistical challenges. Our interdisciplinary approach involves developing novel machine learning methods including to address the statistical and computational challenges inherent to single-cell and imaging data, in particular in clinical samples.
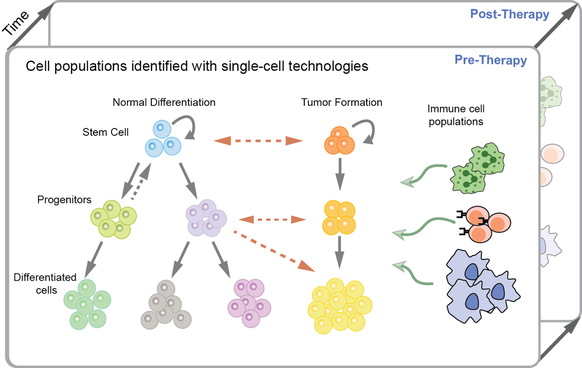
By integrating single-cell multi-omics and imaging data using probabilistic modeling and deep learning approaches, we aim to infer spatiotemporal dynamics, intercellular interactions, and dysregulated programs in the tumor microenvironment. Our statistical models reveal cellular and molecular mechanisms underlying response or resistance to immunotherapies. We also develop models for comparing cell state dynamics between disease and healthy context to pinpoint diverging cell states in particular in the process of tumor initiation.
Ultimately, these interpretable frameworks applied to various cancer systems can guide and improve therapies, predictive biomarkers, and early-detection strategies by accounting for diversity and plasticity of cells.
By integrating single-cell multi-omics and imaging data using probabilistic modeling and deep learning approaches, we aim to infer spatiotemporal dynamics, intercellular interactions, and dysregulated programs in the tumor microenvironment. Our statistical models reveal cellular and molecular mechanisms underlying response or resistance to immunotherapies. We also develop models for comparing cell state dynamics between disease and healthy context to pinpoint diverging cell states in particular in the process of tumor initiation.
Ultimately, these interpretable frameworks applied to various cancer systems can guide and improve therapies, predictive biomarkers, and early-detection strategies by accounting for diversity and plasticity of cells.