Starfysh
|
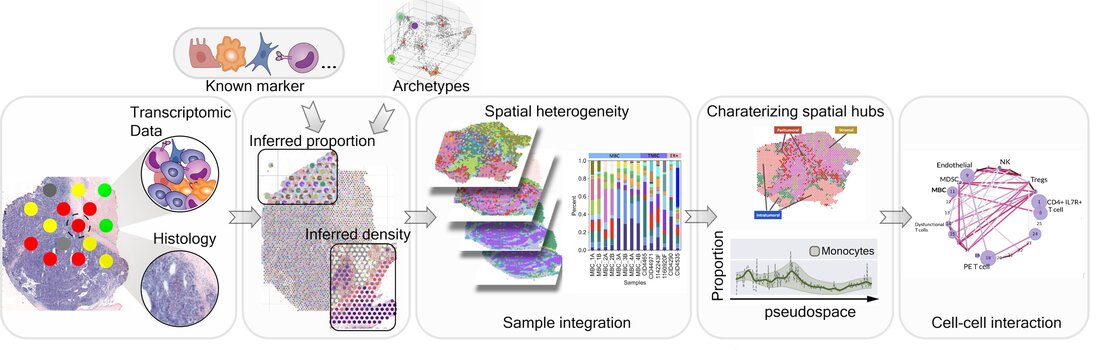
Starfysh (Spatial Transcriptomic Analysis using Reference-Free auxiliarY deep generative modeling and Shared Histology) is an end-to-end toolbox for the analysis and integration of Spatial Transcriptomic (ST) datasets. In summary, the Starfysh framework enables reference-free deconvolution of cell types and refined cell states and can be improved with the integration of paired histology images of tissues, if available. Starfysh is capable of integrating data from multiple tissues. In particular, Starfysh identifies common or sample-specific spatial “hubs” with unique composition of cell types. To uncover mechanisms underlying local and long-range communication, Starfysh can be used to perform downstream analysis on the spatial organization of hubs.
(preprint, highlights, code) |
Decipher
|
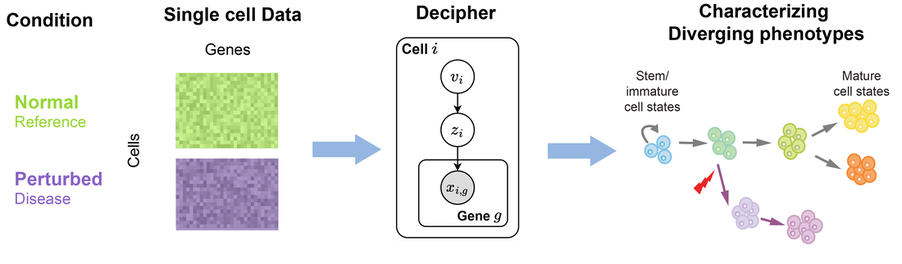
Decipher (DEep CharacterIzation of PHenotypic dERailment) integrates normal and perturbed (e.g. disease) single-cell RNA-seq datasets, to characterize derailed trajectories and reveal shared and disrupted dynamics. The model combines variational autoencoders with deep exponential families to reconstruct trajectories, and allows for correlated latent factors. It obtains a joint pseudotime between trajectories and quantifies disruption in gene expression using a basis decomposition framework.
Decipher further introduces a novel approach to visualize data, without the need for methods such as UMAP or TSNE. The hierarchical (two-tier) dimensionality reduction, which captures complex relationships, can in fact be used to visualize any single-cell dataset without distorting the global geometry. (preprint, highlights, code) |
DIISCO
|
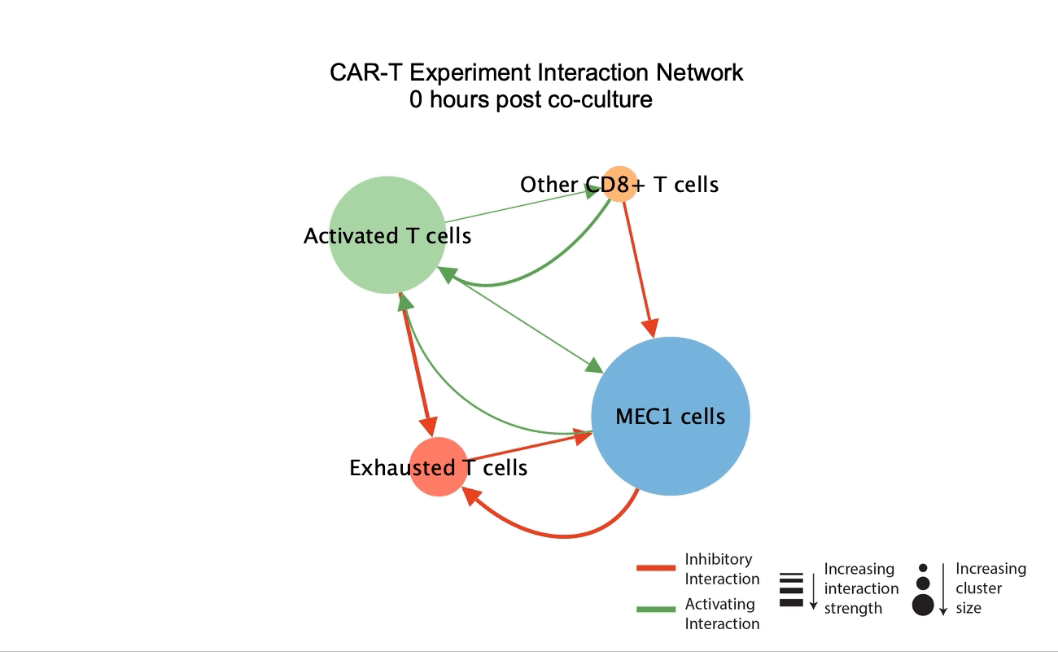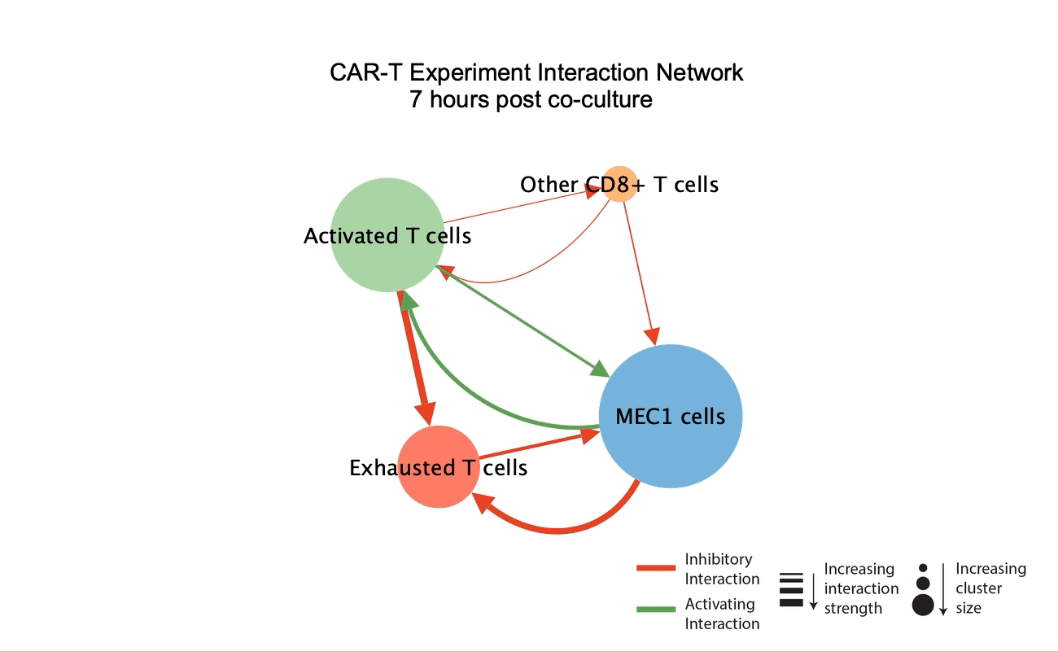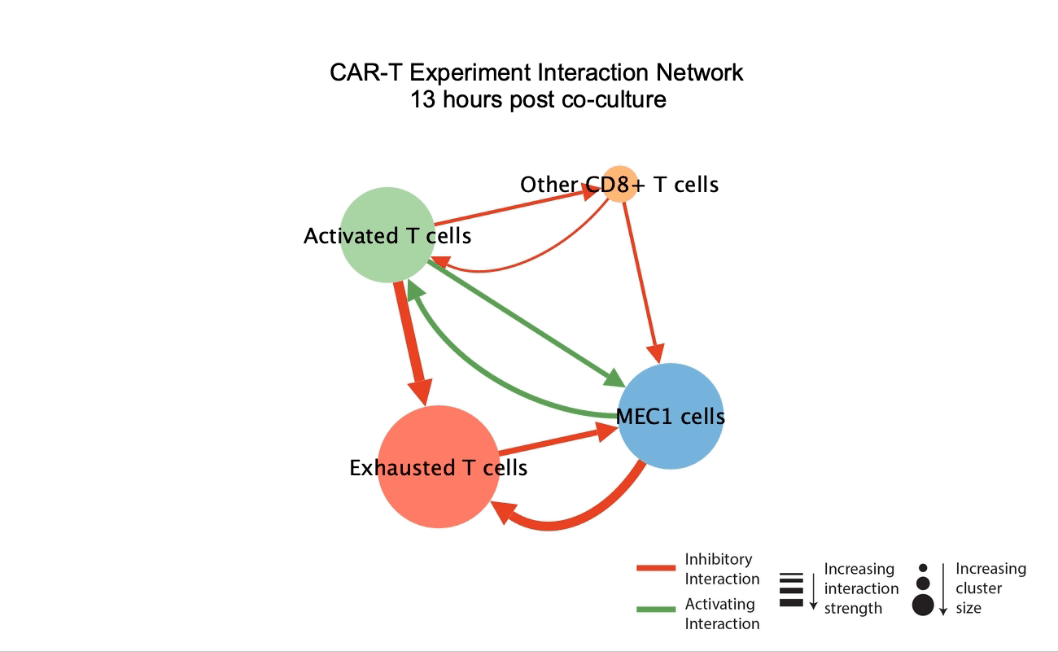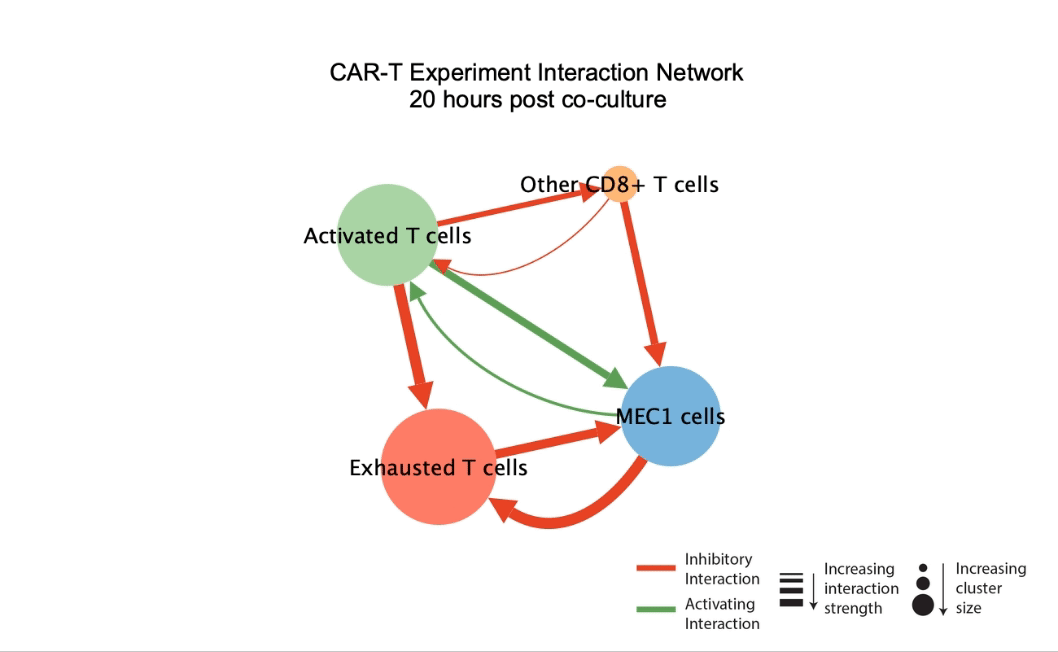
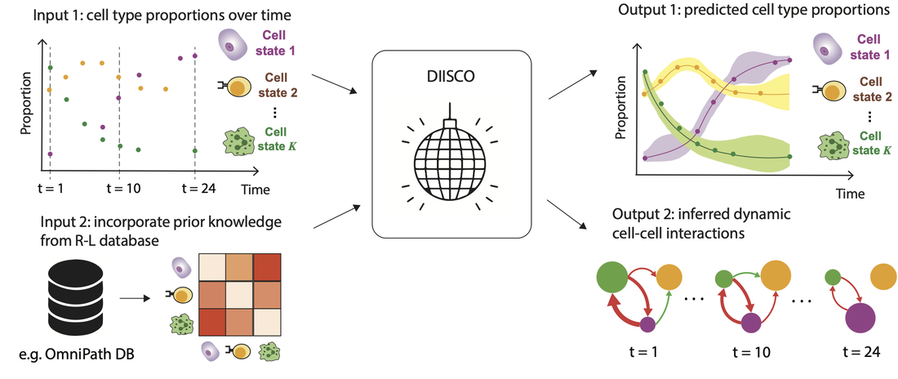
DIISCO (Dynamic Intercellular Interactions in Single Cell transcriptOmics) is a Bayesian framework for characterizing the temporal dynamics of cellular interactions using single-cell RNA-sequencing data from multiple time points. The method uses structured Gaussian process regression to unveil time-resolved interactions among diverse cell types according to their co-evolution and incorporates prior knowledge of receptor-ligand complexes. (preprint, highlights, code) |
SDCD
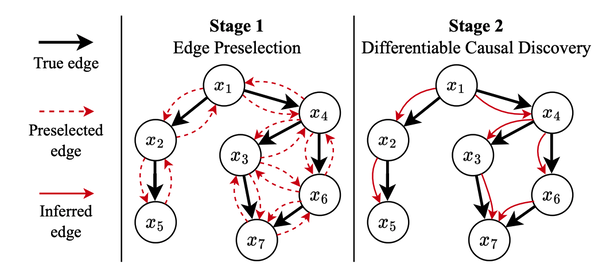
SDCD (Stable Differentiable Causal Discovery) is a method for inferring causal graphs from labeled interventional data. It consists of a two-stage optimization procedure for causal discovery that is stable and computationally efficient. It removes key barriers that previously limited differentiable causal discovery to small problem sizes and is applicable to the size of typical biological datasets, e.g. for gene regulatory network inference.
(preprint, code)
(preprint, code)